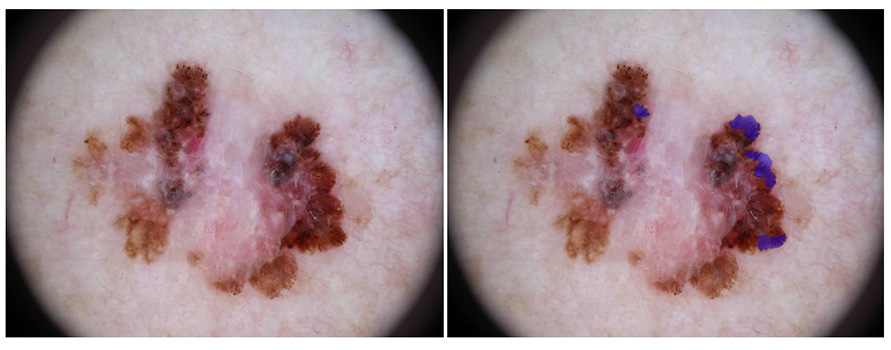
Streaks
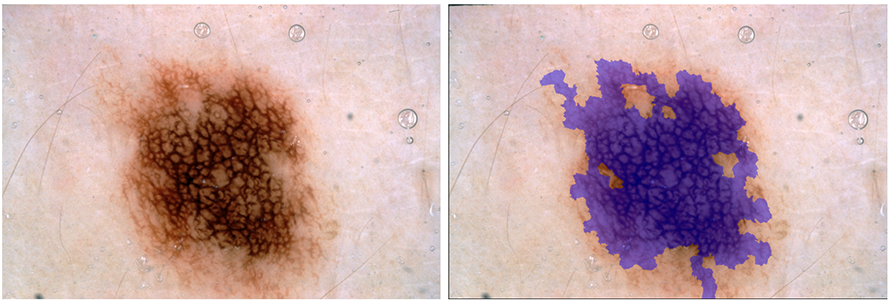
Pigment Network
Goal
Participants are asked to submit automated predictions of clinical dermoscopic features on supplied superpixel tiles. Clinical dermoscopic features are recognized by expert dermatologists when evaluating skin lesions and inform their decision to biopsy suspicious skin lesions.
Data
Lesion dermoscopic feature data includes the original lesion image, a corresponding superpixel mask, and superpixel-mapped expert annotations of the presence and absence of the following features:
- Network
- Negative Network
- Streaks
- Milia-like Cysts
Superpixel Parsing Overview
To reduce the variability and dimensionality of spatial feature annotations, the lesion images have been subdivided into superpixels using the SLIC0 algorithm.
Tools to automatically extract super pixels from images, to help fascinate ingestion into established image classification pipelines, have been developed to assist participants. These tools generate a single image per superpixel, and a corresponding ground truth file, in a similar format to Part 3 of the challenge. The tools are available here.
A lesion image's superpixels should be semantically considered as an integer-valued label map mask image. All superpixel mask images will have the exact same X and Y spatial dimensions as their corresponding lesion image. However, to simplify storage and distribution, superpixel masks are encoded as 8-bit-per-channel 3-channel RGB PNG images. To decode a PNG superpixel image into a label map, use the following algorithm (expressed as pseudocode):
uint32 decodeSuperpixelIndex(uint8 pixelValue[3]) {
uint8 red = pixelValue[0]
uint8 green = pixelValue[1]
uint8 blue = pixelValue[2]
// "<<" is the bit-shift operator
uint32 index = (red) + (green << 8) + (blue << 16)
return index
}
Training Data
Dermoscopy Image & Superpixel Data
The training image and superpixel data file is a ZIP
file, containing 2000 lesion images in JPEG format and 2000
corresponding superpixel masks in PNG format. All lesion
images are named using the scheme
ISIC_<image_id>.jpg, where
<image_id> is a 7-digit unique
identifier. EXIF tags in the lesion images have been
removed; any remaining EXIF tags should not be relied upon
to provide accurate metadata. All superpixel masks are
named using the scheme
ISIC_<image_id>_superpixels.png, where
<image_id> matches the corresponding
lesion image for the superpixel mask.
Ground Truth Annotations
The training ground truth annotations file is a ZIP
file, containing 2000 dermoscopic feature files in JSON
format. All feature files
are named using the scheme
ISIC_<image_id>.json, where
<image_id> matches the corresponding
Training Data lesion image and superpixel mask for the
feature file.
Each feature file contains a top-level JSON Object (key-value map) with 4 keys: "network," "negative network," "streaks," and "milia-like cysts," representing the dermoscopic features of interest. The value of each of theses Object elements is a JSON Array, of length N, where N is the total number of superpixels in the corresponding superpixel mask. Each value within the Array at position k, where 0<= k < N, corresponds to the region within the decoded superpixel index k. The Array values are each JSON Numbers, and equal to either:
0: representing the absence of a given dermoscopic feature somewhere within the corresponding superpixel's spatial extent1: representing the presence of a given dermoscopic feature somewhere within the corresponding superpixel's spatial extent
For example, the feature file:
{
"network": [0, 0, 1, 0, 1, 0],
"streaks": [1, 1, 0, 0, 0, 0]
}
would correspond to a superpixel file with 6 superpixels
(encoded in PNG as R=0, G=0, B=0 through
R=5, G=0, B=0). The lesion image pixels
overlaid by superpixels 2 and 4 (counting from 0) would
contain the "network" dermoscopic feature, while the lesion
image pixels overlaid by superpixels 0 and 1 would contain
the "streaks" dermoscopic feature.
Feature data were obtained from expert superpixel-level annotations, with cross-validation from multiple evaluators.
The dermoscopic features are not mutually exclusive (i.e. both may be present within the same spatial region or superpixel). Any superpixel tile not annotated as positive for a feature may be considered to be a negative example.
Participants are not strictly required to utilize the training data in the development of their lesion classification algorithm and are free to train their algorithm using external data sources. Any such sources must be properly cited in the supplied 4 page abstract.
Submission Instructions
The Test Data files are in a ZIP container, and are the exact same format as the Training Data. The Test Data files should be downloaded via the "Download test dataset" button below. Note: you must be signed-in and registered to participate in this phase of the challenge in order for this link to be visible.
The submitted *Test Results file should be in the same format as the Training Ground Truth file. Specifically, the results file should be a ZIP file of 600 feature files in JSON format. Each feature file should contain the participant's best attempt at a fully automated per-superpixel detection of the features on the corresponding lesion image and superpixel mask. Each feature file should be named and encoded according to the conventions of the training ground truth.
Note, the JSON Numbers in the submitted Test Results
should not be only 0.0 and
1.0, but instead should be floating-point
values in the closed interval [0.0, 1.0],
where values:
0.0to0.5: represent some confidence that the feature is absent from the lesion image anywhere within the given superpixel, with relatively lesser values indicating relatively more confidence in the absence> 0.5to1.0: represent some confidence that the feature is present in the lesion image anywhere within the given superpixel, with relatively greater values indicating relatively more confidence in the presence
Note, arbitrary score ranges and thresholds can be converted to the range of 0.0 to 1.0, with a threshold of 0.5, trivially using the following sigmoid conversion:
1 / (1 + e^(-(a(x - b))))
where x is the original score,
b is the binary threshold, and
a is a scaling parameter (i.e. the inverse
measured standard deviation on a held-out dataset).
Evaluation
Participants will be ranked and awards granted based only on AUC. However, submissions will additionally be compared using using a variety of common classification metrics, including:
Some useful resources for metrics computation include:
