Goal
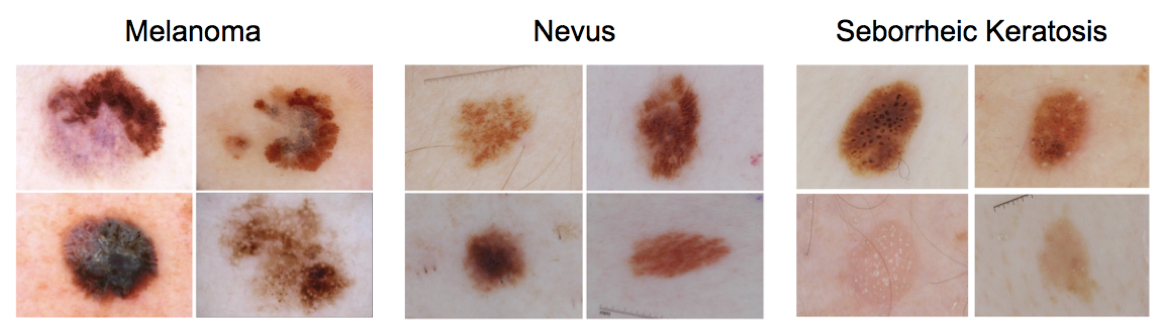
In this task, participants are asked to complete two independent binary image classification tasks that involve three unique diagnoses of skin lesions (melanoma, nevus, and seborrheic keratosis). In the first binary classification task, participants are asked to distinguish between (a) melanoma and (b) nevus and seborrheic keratosis. In the second binary classification task, participants are asked to distinguish between (a) seborrheic keratosis and (b) nevus and melanoma.
Definitions:
- Melanoma – malignant skin tumor, derived from melanocytes (melanocytic)
- Nevus – benign skin tumor, derived from melanocytes (melanocytic)
- Seborrheic keratosis – benign skin tumor, derived from keratinocytes (non-melanocytic)
Data
Lesion classification data includes the original image, paired with a gold standard (definitive) diagnosis, referred to as "Ground Truth".
Training Image Data
2000 images are provided as training data, including 374 "melanoma", 254 "seborrheic keratosis", and the remainder as benign nevi (1372). The training data is provided as a ZIP file, containing dermoscopic lesion images in JPEG format and a CSV file with some clinical metadata for each image.
All images are named using the scheme
ISIC_<image_id>.jpg, where
<image_id> is a 7-digit unique
identifier. EXIF tags in the images have been removed; any
remaining EXIF tags should not be relied upon to provide
accurate metadata.
The CSV file contains three columns:
image_id, identifying the image that the row corresponds toage_approximate, containing the age of the lesion patient, rounded to 5 year intervals, or"unknown"sex, containing the sex of the lesion patient, or"unknown"
Ground Truth Data
The Training Ground Truth file is a single CSV (comma-separated value) file, containing 3 columns:
- The first column of each row contains a string of
the form
ISIC_<image_id>, where<image_id>matches the corresponding Training Data image. - The second column of each row pertains to the first
binary classification task (melanoma vs. nevus and
seborrheic keratosis) and contains the value 0 or 1.
- The number 1 = lesion is melanoma
- The number 0 = lesion is nevus or seborrheic keratosis
- The third column of each row pertains to the second
classification task (seborrheic keratosis vs. melanoma
and nevus) and contains the value 0 or 1.
- The number 1 = lesion is seborrheic keratosis
- The number 0 = lesion is melanoma or nevus
Malignancy diagnosis data were obtained from expert consensus and pathology report information. Participants are not strictly required to limit development to the training data, and are free to train their algorithm using external data sources. However, any other sources of data in system development must be properly cited in the abstract.
Submission Instructions
The Test Data files are in a ZIP container, and are the exact same format as the Training Data. The Test Data files should be downloaded via the "Download test dataset" button below. Note: you must be signed-in and registered to participate in this phase of the challenge in order for this link to be visible.
The submitted Test Results file should use the same format as the Training
Ground Truth file. The first column of each row should contain a string of the form
ISIC_<image_id>, where <image_id> matches a
corresponding Test Data image. The second and third column of each row
should contain a floating-point value in the closed interval [0.0, 1.0], where
0.5 is used as the binary classification threshold.
The second column of each row should pertain to the first binary classification task (melanoma vs. nevus and seborrheic keratosis). The third column of each row should pertain to the second binary classification task (seborrheic keratosis vs. melanoma and nevus).
Note that arbitrary score ranges and thresholds can be converted to the range of 0.0 to 1.0, with a threshold of 0.5, trivially using the following sigmoid conversion:
1 / (1 + e^(-(a(x - b))))
where x is the original score, b is the binary threshold, and
a is a scaling parameter (i.e. the inverse measured standard deviation on a
held-out dataset). Participants are asked to set their binary threshold 'b' to a value where
the classification system is expected to achieve 89% sensitivity, although this is not
required.
Evaluation
Participants will be ranked according to each category individually, as well as the average performance across both categories (giving rise to the possibility of 3 distinct "winners"). Ranks and awards will be assigned based only on area under the receiver operating characteristic curve (AUC). However, submissions will also be evaluated using using a variety of common binary classification metrics, reported for scientific completeness, including:
- sensitivity at 0.5 confidence threshold
- specificity at 0.5 confidence threshold
- accuracy at 0.5 confidence threshold
- average precision evaluated at sensitivity of 100%
- specificity evaluated at a sensitivity of 82%
- specificity evaluated at a sensitivity of 89%
- specificity evaluated at a sensitivity of 95%
- area under the receiver operating characteristic curve (AUC)
Some useful resources for metrics computation include:
