Goal
Submit automated predictions of the locations of dermoscopic attributes (established clinically-meaningful visual skin lesion patterns) within dermoscopic images.
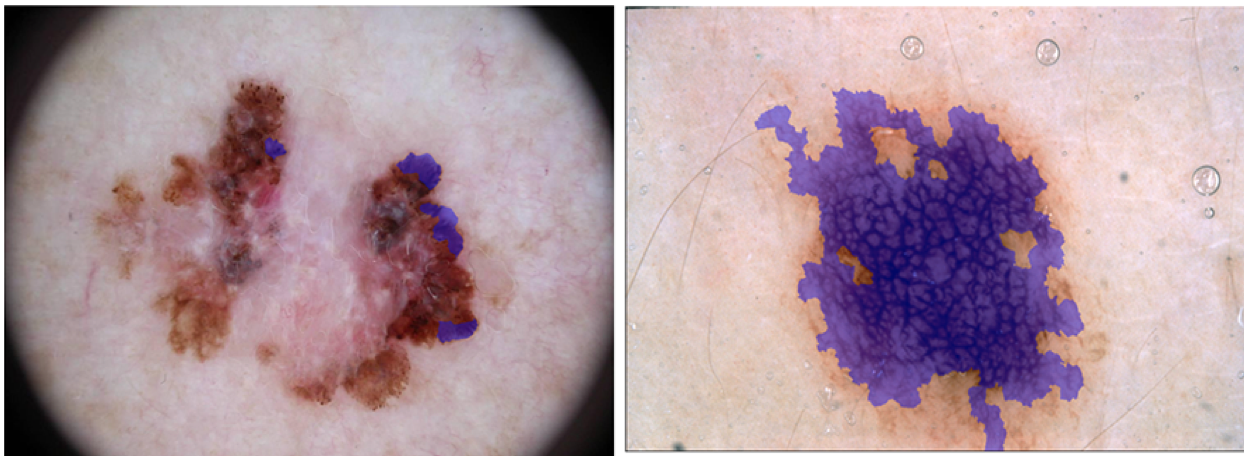
The following dermoscopic attributes should be identified:
- pigment network
- negative network
- streaks
- milia-like cysts
- globules (including dots)
Data
Input Data
The input data are dermoscopic lesion images in JPEG format.
All lesion images are named using the scheme ISIC_<image_id>.jpg,
where <image_id> is a 7-digit unique identifier. EXIF tags in the
images have been removed; any remaining EXIF tags should not be relied upon to provide
accurate metadata.
The lesion images were acquired with a variety of dermatoscope types, from all anatomic sites (excluding mucosa and nails), from a historical sample of patients presented for skin cancer screening, from several different institutions. Every lesion image contains exactly one primary lesion; other fiducial markers, smaller secondary lesions, or other pigmented regions may be neglected.
The distribution of disease states represent a modified "real world" setting whereby there are more benign lesions than malignant lesions, but an over-representation of malignancies.
Response Data
The response data are binary mask images in PNG format, indicating the location of a dermoscopic attribute within each input lesion image.
Mask images are named using the scheme ISIC_<image_id>_attribute_<attribute_name>.png, where
<image_id> matches the corresponding lesion image for the mask
and <attribute_name> identifies a dermoscopic attribute:
pigment_networknegative_networkstreaksmilia_like_cystglobules
Mask images must have the exact same dimensions as their corresponding lesion image. Mask images are encoded as single-channel (grayscale) 8-bit PNGs (to provide lossless compression), where each pixel is either:
0: indicating areas where the dermoscopic attribute is absent255: indicating areas where the the dermoscopic attribute is present
Regions with a given attribute are not necessarily contiguous within an image.
Ground Truth Provenance
Mask image ground truth (provided for training and used internally for scoring validation and test phases) data were generated by expert dermatology research fellows, supervised by a recognized dermoscopy expert, via a manual image annotation process of selecting SLIC superpixels for each attribute within each image.
Evaluation
Goal Metric
Predicted responses are scored using the Jaccard index. To compute, images are standardized to a fixed size (i.e. 256x256). Then all prediction pixels across the entire dataset (not image-by-image, as some images may have no positive instances of a dermascopic attribute) are compared to ground truth pixels.
Other Metrics
Participants will be ranked and awards granted based only on the Jaccard index. However, for scientific completeness, predicted responses will also have the following metrics computed on a pixel-wise basis (comparing prediction vs. ground truth) for each attribute in each image:
Submission Instructions
To participate in this task:
- Train
- Download the training input data and training ground truth response data.
- Develop an algorithm for locating dermoscopic attributes in general.
- Validate (optional)
- Download the validation input data.
- Run your algorithm on the validation Input data to produce validation predicted responses.
- Submit these validation predicted responses to receive an immediate score. This will provide feedback that your predicted responses have the correct data format and have reasonable performance. You may make unlimited submissions.
- Test
- Download the test input data.
- Run your algorithm on the test input data to produce test predicted responses.
- Submit these test predicted responses. You may submit a maximum of 3 separate approaches/algorithms to be evaluated independently. You may make unlimited submissions, but only the most recent submission for each approach will be used for official judging. Use the "brief description of your algorithm’s approach" field on the submission form to distinguish different approaches. Previously submitted approaches are available in the dropdown menu.
- Submit a manuscript describing your algorithm’s approach.
