Goal
Submit automated predictions of disease classification within dermoscopic images.
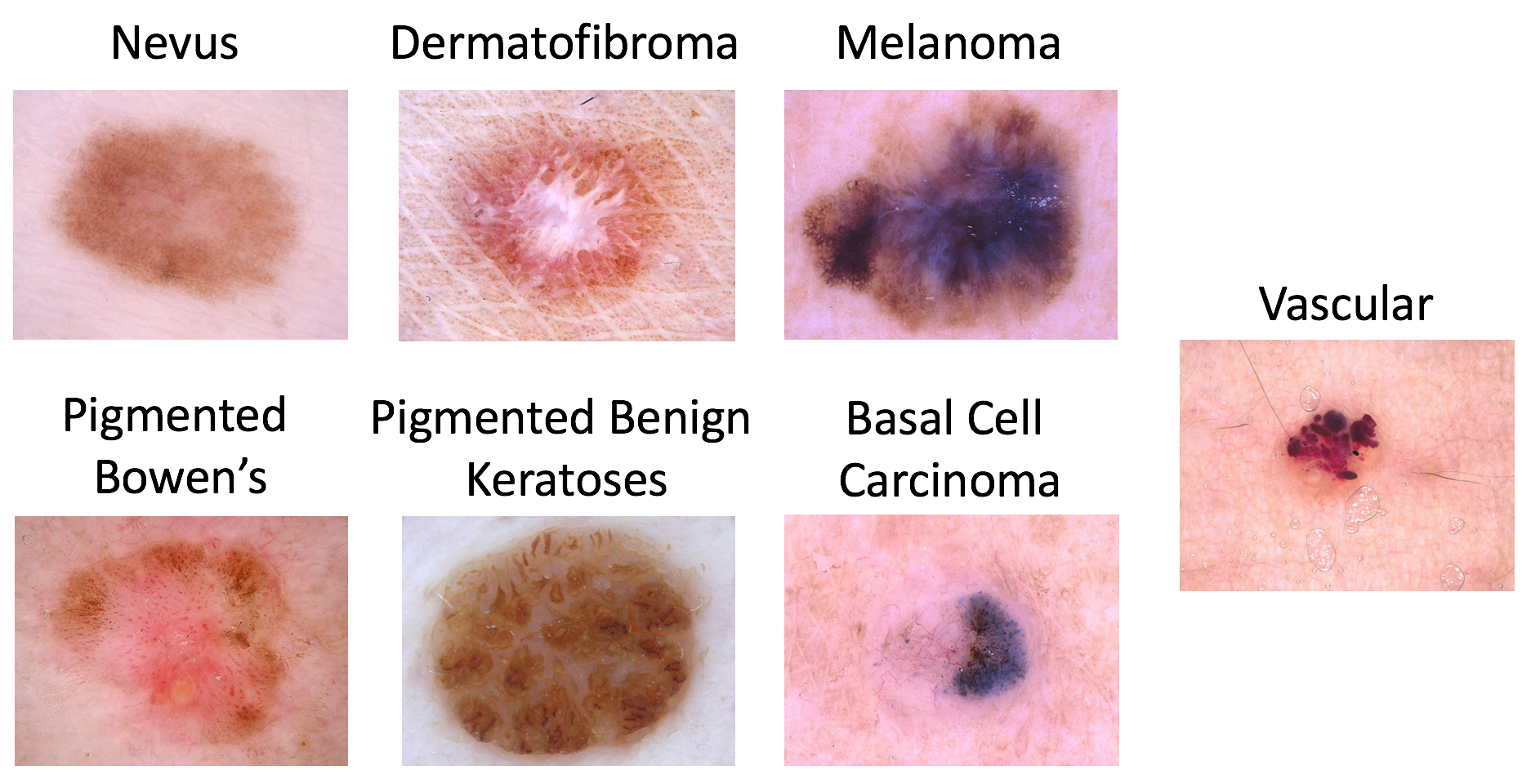
Possible disease categories are:
- Melanoma
- Melanocytic nevus
- Basal cell carcinoma
- Actinic keratosis / Bowen’s disease (intraepithelial carcinoma)
- Benign keratosis (solar lentigo / seborrheic keratosis / lichen planus-like keratosis)
- Dermatofibroma
- Vascular lesion
Data
Input Data
The input data are dermoscopic lesion images in JPEG format.
All lesion images are named using the scheme ISIC_
, where
is a 7-digit unique identifier. EXIF tags in the images have been removed; any
remaining EXIF tags should not be relied upon to provide accurate metadata.
The lesion images come from the HAM10000 Dataset, and were acquired with a variety of dermatoscope types, from all anatomic sites (excluding mucosa and nails), from a historical sample of patients presented for skin cancer screening, from several different institutions. Images were collected with approval of the Ethics Review Committee of University of Queensland (Protocol-No. 2017001223) and Medical University of Vienna (Protocol-No. 1804/2017).
The distribution of disease states represent a modified "real world" setting whereby there are more benign lesions than malignant lesions, but an over-representation of malignancies.
Response Data
The response data are sets of binary classifications for each of the 7 disease states, indicating the diagnosis of each input lesion image.
Response data are all encoded within a single CSV file (comma-separated value) file, with each classification response in a row. File columns must be:
image: an input image identifier of the formISIC_MEL: "Melanoma" diagnosis confidenceNV: "Melanocytic nevus" diagnosis confidenceBCC: "Basal cell carcinoma" diagnosis confidenceAKIEC: "Actinic keratosis / Bowen’s disease (intraepithelial carcinoma)" diagnosis confidenceBKL: "Benign keratosis (solar lentigo / seborrheic keratosis / lichen planus-like keratosis)" diagnosis confidenceDF: "Dermatofibroma" diagnosis confidenceVASC: "Vascular lesion" diagnosis confidence
Diagnosis confidences are expressed as floating-point values in the closed interval [0.0,
1.0], where 0.5 is used as the binary classification threshold. Note
that arbitrary score ranges and thresholds can be converted to the range of 0.0
to 1.0, with a threshold of 0.5, trivially using the following
sigmoid conversion:
1 / (1 + e^(-(a(x - b))))where x is the original score, b is the binary threshold, and
a is a scaling parameter (i.e. the inverse measured standard deviation on a
held-out dataset). Predicted responses should set the binary threshold b to a
value where the classification system is expected to achieve 89% sensitivity, although this
is not required.
Predicted diagnosis confidence values may vary independently, though exactly one disease state is actually present in each input lesion image.
Ground Truth Provenance
As detailed in the HAM10000 Dataset description, diagnosis ground truth (provided for training and used internally for scoring validation and test phases) were established by one of the following methods:
- Histopathology
- Reflectance confocal microscopy
- Lesion did not change during digital dermatoscopic follow up over two years with at least three images
- Consensus of at least three expert dermatologists from a single image
In all cases of malignancy, disease diagnoses were histopathologically confirmed.
Evaluation
Goal Metric
Predicted responses are scored using a normalized multi-class accuracy metric (balanced across categories). Tied positions will be broken using the area under the receiver operating characteristic curve (AUC) metric.
Rationale
Clinical application on skin lesion classification has two goals eventually: Giving specific information and treatment options for a lesion, and detecting skin cancer with a reasonable sensitivity and specificity. The first task needs a correct specific diagnosis out of multiple classes, whereas the second demands a binary decision "biopsy" versus "don’t biopsy". In the former ISIC challenges, focus was on the second task, therefore this year we want to rank for the more ambitious metric of normalized multiclass accuracy, as it is also closer to real evaluation of a dermatologist. This is also important for the extending reader study, where the winning algorithm(s) will be compared to physicians performance in classification of digital images.
Other Metrics
Participants will be ranked and awards granted based only on the multiclass accuracy metric. However, for scientific completeness, predicted responses will also have the following metrics computed (comparing prediction vs. ground truth) for each image:
Individual Category Metrics
- sensitivity
- specificity
- accuracy
- area under the receiver operating characteristic curve (AUC)
- mean average precision
- F1 score
- AUC integrated between 80% to 100% sensitivity (AUC80) for Melanoma diagnosis only
- positive predictive value (PPV)
- negative predictive value (NPV)
Aggregate Metrics
- average AUC across all diagnoses
- malignant vs. benign diagnoses category AUC
Submission Instructions
To participate in this task:
- Train
- Download the training input data and training ground truth response data.
- Develop an algorithm for generating lesion diagnosis classifications in general.
- Validate (optional)
- Download the validation input data.
- Run your algorithm on the validation Input data to produce validation predicted responses.
- Submit these validation predicted responses to receive an immediate score. This will provide feedback that your predicted responses have the correct data format and have reasonable performance. You may make unlimited submissions.
- Test
- Download the test input data.
- Run your algorithm on the test input data to produce test predicted responses.
- Submit these test predicted responses. You may submit a maximum of 3 separate approaches/algorithms to be evaluated independently. You may make unlimited submissions, but only the most recent submission for each approach will be used for official judging. Use the "brief description of your algorithm’s approach" field on the submission form to distinguish different approaches. Previously submitted approaches are available in the dropdown menu.
- Submit a manuscript describing your algorithm’s approach.
